A multi-modal data resource for investigating geographic heterogeneity in patient-derived xenograft tumors
Licensed according to this deed.
Category
Published on
Abstract
PSON0010 Cancer Sampling Index – CaSIX
Study Contact: Lani Wu lani.wu@ucsf.edu
Download the dataset at ftp://caftpd.nci.nih.gov/psondcc/casix/
Studies using Patient Derived Xenograft (PDX) models of human cancer permit a wider range of experimental approaches to measure and map tumor heterogeneity than what would be possible in a patient sample. The CaSIX project seeks to generate a dataset to use as a basis for providing a spatial delineation of heterogeneity in tumors at the cellular and molecular level. This assessment entails genomic, transcriptional and proteomic data and physical characterization across multiple sites within an individual tumor.
This was investigated in the context of four colon-cancer PDX models ( two each from the left and right colon) whose biological properties were assayed using genetic, transcriptomic, proteomic and immunofluorescence microscopy based assays.
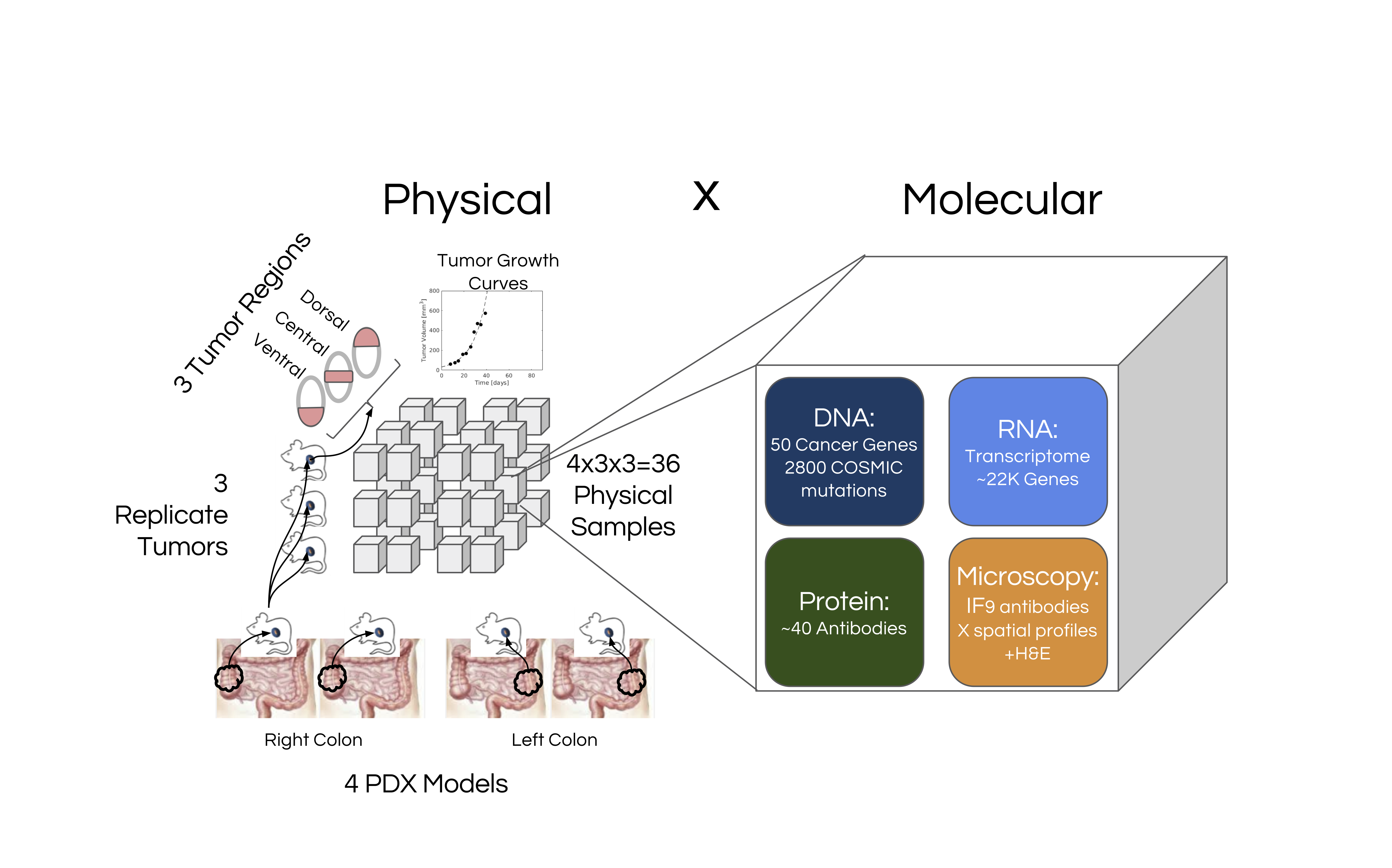
Figure 1. Overview of the CaSix project
For each of the PDX models, 3 replicate tumors were grown, and each replicate tumor was divided into three regions (dorsal, central and ventral). Thus in total 36 = 4×3×3 PDX samples were generated, and each was profiled generating the datasets as follows:
Growth_Curves- This dataset represents the growth and harvesting of the PDX tumors stored as xls files.
Microscopy H & E – Brightfield microscopy images of hematoxylin and eosin (h&e) stained histological section extracted from the PDX samples were captured.
Microscopy_IF – Immunofluorescence (IF) microscopy images of histological section extracted from the PDX samples. Immunofluorescence microscopy involved staining each section for four biomarkers and imaging with an Aperio Fluorescence Digital Scanner. Accordingly, the imaging can be thought of as being divided into 4 marker sets of DAPI, vimentin and the markers for the following proteins:
- betacatenin, rps6
- ki67, erk
- ecadherin, stat3
- akt, vegf
DNA – Ion Torrent technology was used to perform targeted sequencing of 50 genes using the Ion Ampliseq Cancer Hotspot Panel v2 kit. The raw data consists of sequenced reads of fastq files. Processed data consists of variant calls in files of vcf format.
RNA – The transcriptome profiling of the PDX samples was done using Ion Torrent sequencing based on Ion Ampliseq Transcriptome Human Gene Expression Kits, targeting > 20,0000 RefSeq transcripts. Raw data consists of fastq files of the sequenced reads. Processed data consists of reads-per-million normalized counts for various genes based on mapping of reads to the human transcriptome and stored as tab-delimited text files and bam files using the set of RefSeq transcripts as reference sequences.
RPPA – The PDX samples underwent proteomic profiling using Reverse Phase Protein Array with 46 antibodies, performed by George Mason University (GMU). Laser capture microdissection (LCM) followed by lysate extraction was performed on all 36 samples to extract lysates for a) just tumor cells b) residuals cells after tumors were removed and c) whole tissue sections. Additionally for 5 samples lysates from stromal cells extracted via LCM was generated, giving a total of 113 lysates which were profiled using RPPA.
A design principle of this study was being able to focus specifically on tumor (as opposed to stromal tissue) properties. For the RNA/DNA/RPPA assays this was achieved by using Laser Capture Microdissection to capture extract and profile tumor regions. For the Microscopy, a stroma specific marker (vimentin) was included in all IF images.
Data usage policy
The data contained within the PS-ON DCC is based on several research projects and is intended to be rapidly and constantly updated for the research community to access and use. The NCI requests that any data users:
Inform the data submitters about the intention to submit a publication that uses PS-ON DCC data.
Include the following statement in any publications resulting from the use of PS-ON DCC data: Data used in this publication were generated by projects sponsored by the NCI Physical Sciences in Oncology Initiative.
Acknowledgments
We wish to thank: Debra Hope, Michael Espy, and Joan Pontius for critical feedback throughout the project; and Emmanuel Petricoin, Mariaelena Pierobon, Elisa Baldelli, Audrey Papp and Amanda Williams for their invaluable support in the profiling assays.
Cite this work
Researchers should cite this work as follows: